Agentic discovery platform
Multi-omics insights, powered by 100% of the genome
Lucid combines proprietary genomic variant detection, Dark Genome expertise, and multi-omics analysis - including transcriptomics and proteomics - to map 3D genome architecture and link regulatory elements to target genes.
We uncover genomics-driven disease mechanisms, enabling novel biomarkers, target discovery, and patient stratification.
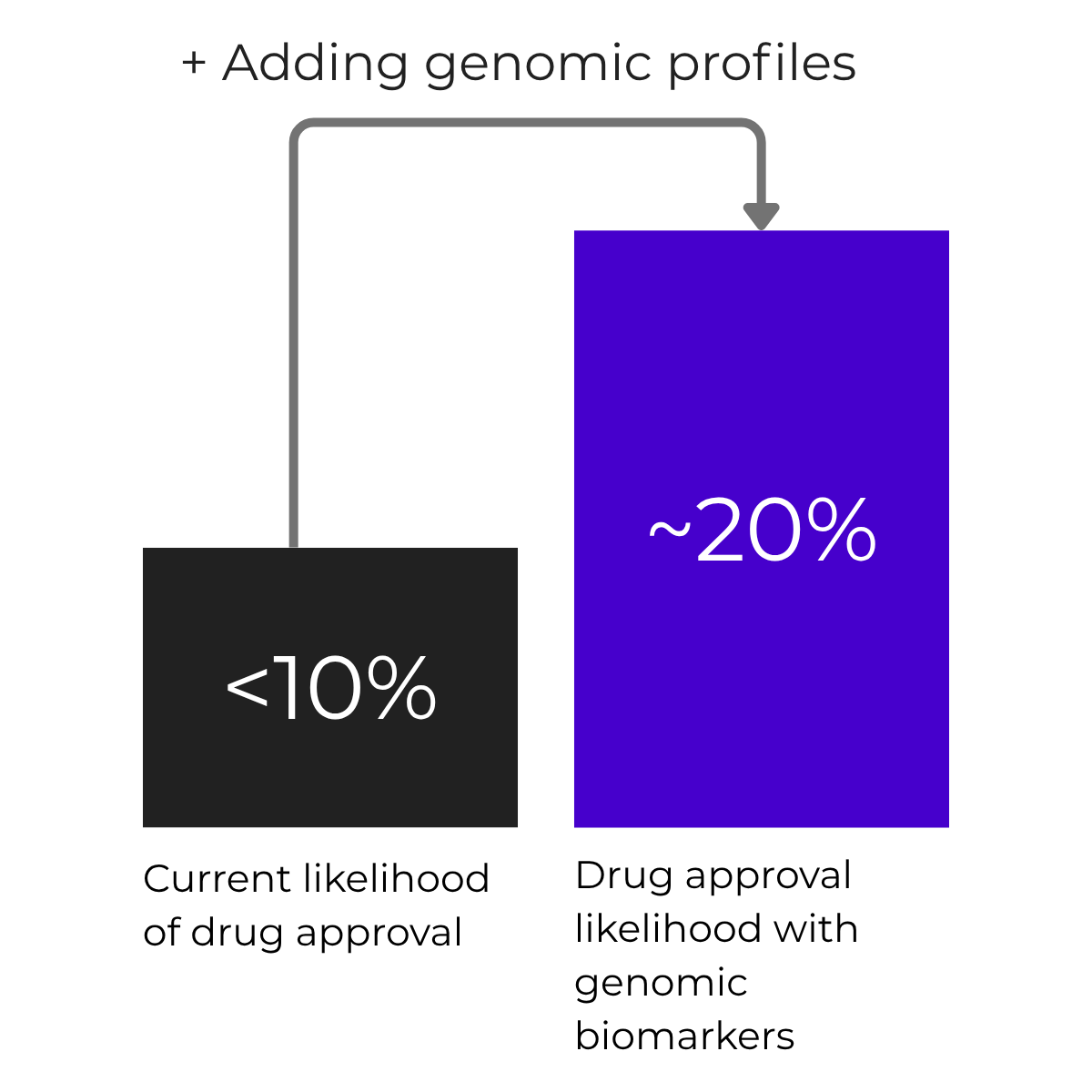
Only 1 in 10 drugs makes it to market
Genomics-guided drugs are 2.6× more likely to reach approval, raising success rates from under 10% to around 20%.
Lucid's team has been at the forefront of non-coding and structural variant research, well before these regions gained wider recognition. We bring deep expertise in uncovering regulatory drivers that conventional pipelines miss.
Why genomics-driven trials?
FASTER & CHEAPER
40%
Reduction in sample size
By including genomic biomarkers in trials while maintaining statistical power
MORE SUCCESSFUL
2.6x
Improved market approval rate
Based on patient stratification and identification of super and non-responders
NEW MARKET OPPORTUNITY
60%
Cancer trials already include genomics
Other therapeutic areas are quickly catching up with oncology in using genomics in trials
Our solutions
Genomics-driven
biomarker discovery
Uncover novel, functional biomarkers including non-coding and structurally complex genomic regions that are inaccessible to standard analytical pipelines.
Leverage freedom to operate in a largely unpatented space of non-coding biomarkers
Uncover disease mechanisms invisible to coding-only approaches (neglecting the Dark Genome)
Identify regulatory elements driving disease initiation, progression and resistance
Genomics-driven
patient stratification
Whole genome analysis of patients to identify potential super and non-responders for clinical trials to adjust sample setup and to boost trial success likelihood
Reduce sample size of clinical studies while maintaining statistical power
Identify Super and Non-responders to increase clinical trial success likelihood
Enable genomic driven adaptive trial arms to reduce trial operations costs
Genomics-driven
Target Identification
Understand complex disease mechanisms and validate novel drug targets by mapping the complete regulatory architecture of the genome.
Understand variant-to-function pathways to prioritize real causal drivers
Discover targets in previously overlooked genomic regions (largely patent free space)
De-risk target selection with stronger genetic evidence (GWAS and functional genomics)
Technology-agnostic analysis
We support all sequencing technologies and apply our proprietary AI data cleaning model to aggregate and filter out background noise.
-
Short-reads
e.g. Illumina
-
Long-reads
e.g. PacBio
-
Linked-reads
e.g TELL-Seq
-
Optical mapping
e.g. Bionano
Multi-omics analysis across
data types
Our platform analyzes data from panels to genomes, providing a manageable, prioritized list of suggested causative pathogenic mutations.
Whole genomes
Gene panels
Exomes
RNA-seq / Transcriptomics
Hi-C
ChIP-seq
Proteomics
EHR data
Case study
A non-coding deleted DNA segment downstream of PITX2 disrupted a TAD boundary element, leading to atrial-specific misexpression and atrial fibrillation. The regulatory mechanism in vivo highlights the impact of non-coding structural variants on cardiac rhythm.
Step 1: Cohort Analysis
Several overlapping structural variants located ~800kb downstream of the PITX2 gene, a known cardiac development factor, were identified in whole-genome sequencing data from atrial fibrillation patients. These variants were absent in SNP-array data.
Step 2: Mechanism of Action
A non-coding CTCF/TAD-boundary deletion fuses adjacent TADs and rewires long-range contacts, leading to PITX2 misexpression
Step 3: In Vivo Validation
The direct causal link between the structural variant and atrial fibrillation susceptibility was subsequently confirmed in a targeted mouse model, validating the finding from discovery to function.
Published in:
Discover our Platform
Get a personalized walkthrough of our platform and see how it can benefit your needs.